wwPDB 2026 News
Contents
02/10/2026
New PDB Beta Archive Available for Testing
By 2028 4-character PDB IDs (e.g. 1abc) will be fully allocated. After that, all new entries will be assigned only extended PDB IDs.
The new extended PDB ID format will be 12 characters, which includes a prefix pdb_ followed by 8 alphanumeric characters, e.g. pdb_1000axyz. This new ID format will enable text mining detection of PDB entries in the published literature and allow for more informative and transparent delivery of revised data files. When submitting extended PDB IDs to journals and citing extended PDB IDs in manuscripts, all 12 characters including prefix pdb_ should be provided.
A PDB Beta Archive is now available to help community adopt extended PDB ID and PDBx/mmCIF format during the transition phase. All files at this archive are re-organized with extended PDB ID (including file naming and directories) at entry level, mirroring the same data organization of the PDB Versioned Archive.
All data files for a particular entry are stored in a single directory, labeled based on a two-character hash generated from the penultimate two characters of the PDB code, i.e., https://files-beta.wwpdb.org/pub/wwpdb/pdb/data/entries/<two-letter-hash>/<pdb_accession_code>/<entry_data_File_names>. The two-letter hash will be based on the second and third characters from the last character. For example, PDB entry pdb_1abc5678 will be under /67/. This will maintain consistency with the current PDB archive: PDB entry 1abc is under /ab.
File naming is standardized such that the file type is used for the extension.
For example, file naming is changed from r116dsf.ent.gz to pdb_0000116d-sf.cif.gz for the structure factor file and from pdb318d.ent.gz to pdb_0000318d.pdb.gz for the legacy PDB formatted coordinate file.
When four character PDB IDs are about to be consumed, this PDB Beta Archive will replace the current PDB Archive (expect to be around mid-2027) and entries with extended PDB IDs issued are not compatible with PDB format. wwPDB encourages scientific journals, PDB community and users to transition to PDBx/mmCIF format and adopt new PDB ID format as earlier as possible.
For any further information please contact us at info@wwpdb.org.
 New PDB beta archive available for testing in support of extended PDB IDs
New PDB beta archive available for testing in support of extended PDB IDs
01/19/2026
EMDB Surpasses 10,000 Entries in a Year
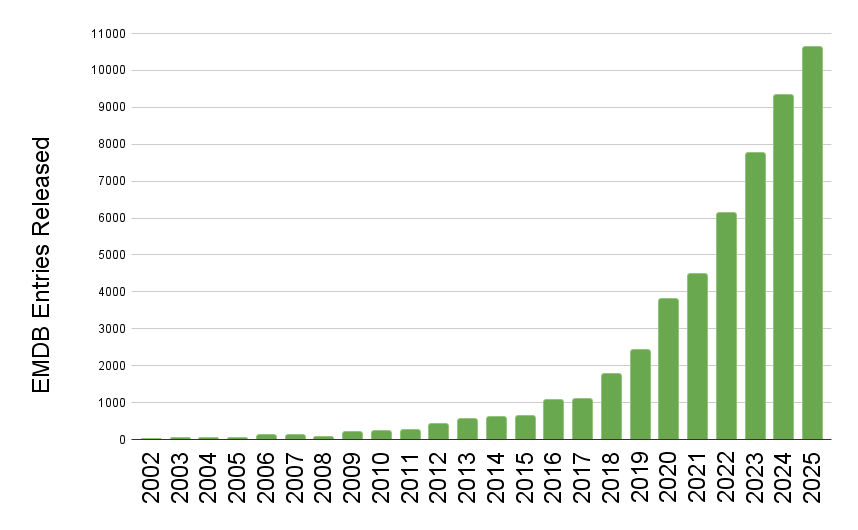 EMDB released entry growth by year
EMDB released entry growth by yearThe wwPDB has reached a historic milestone, releasing 10,000 EMDB entries in a single year.
The rate of release of 3DEM data to the public continues at an extraordinary pace, for the first time the wwPDB has seen the release of 10,000 entries from the EMDB in just a single year. To put this in context, in 2019 the EMDB stood at 10,000 entries in total, after 17 years of growth. Now in 2025 due to the current volume of data release, the archive has surpassed 50,000 total entries.
This rapid growth is a testament to the hard work of the community which the wwPDB endeavors to support. It also highlights the ongoing challenge the community is facing with ever more data created requiring streamlined deposition, processing, curation, peer-review, and sustainable & scalable archiving practices. We are excited to continue to be part of structural biology history.
01/12/2026
wwPDB Shared Responsibilities on Data Processing
Depositions are automatically distributed to wwPDB deposition centers geographically.
Data processing responsibilities are shared among wwPDB partner sites:
- RCSB PDB is responsible for PDB and EMDB data coming from the Americas
- PDBe and EMDB are responsible for PDB and EMDB data from Europe (except Belarus and Russian Federation) and Africa
- PDBj is responsible for PDB and EMDB data from Asia regions (except China), Belarus, Russian Federation and Oceania
- PDBc is responsible for PDB and EMDB data from China
- BMRB is responsible for BMRB data from the Americas, Africa, and Europe
- BMRBj is responsible for BMRB data from Asia
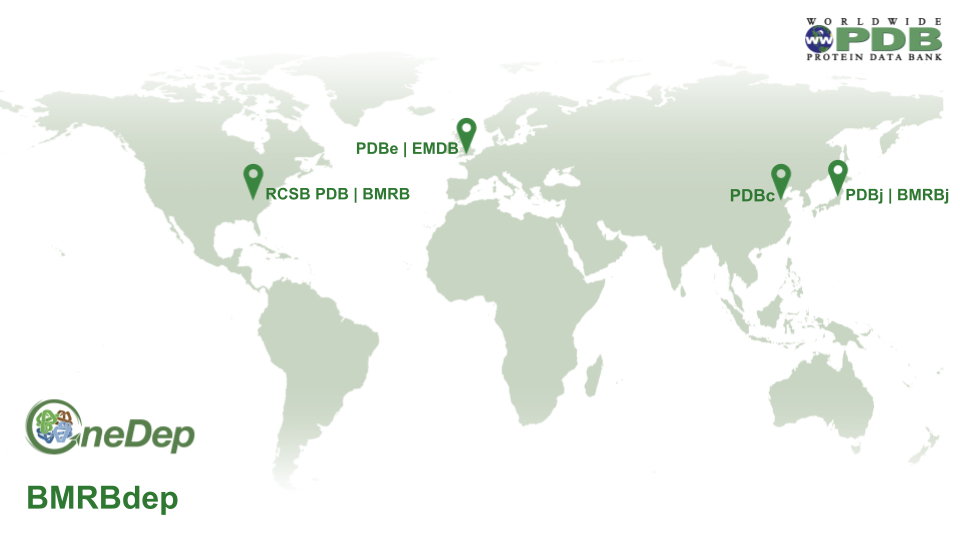 Data processing distribution map
Data processing distribution map
01/11/2026
Register for PDB Workshops at IUCr2026
wwPDB will be part of the Twenty-Seventh Congress and General Assembly of the International Union of Crystallography (IUCr2026) that will be held August 11-18, 2026 in Calgary, Alberta (Canada).
Register now to join the wwPDB for free, half-day and hands-on workshops that will be held just before the Congress begins.
Enhancing PDB Deposition and Validation Practices (afternoon session on Monday, August 10, 2026)
 Pre-meeting workshop: OneDep
Pre-meeting workshop: OneDepThis workshop will provide participants with essential knowledge and skills to easily prepare PDB depositions, evaluate structure quality, and utilize OneDep features to streamline and optimize deposition processes.
Through short lectures and hands-on practical sessions, participants will gain proficiency in making successful PDB depositions using available tools for preparation of necessary data and metadata files consistent with OneDep requirements of the wwPDB OneDep system for complete deposition, rigorous validation, and expert biocuration. Participants will develop a comprehensive understanding of information presented within wwPDB validation reports and the importance in assessing data quality. Practical insights will be shared, enabling participants to confidently undertake and complete successful PDB submissions.
Workshop attendees will also become proficient in leveraging various features and tools integrated into the structure determination software packages and OneDep software system, including automatic initiation of new depositions using application programming interface (API) or using traditional web interface via ORCiD to create deposition sessions, and validating deposited data.
Enabling PDB Deposition of Integrative/Hybrid Methods Structures Determined using 3D Electron Microscopy plus Chemical Crosslinking and Other Complementary Approaches with the PDB-IHM System (afternoon session on Tuesday, August 11, 2026)
 Pre-meeting workshop: IHM Deposition
Pre-meeting workshop: IHM DepositionThis workshop will enable structural biologists to learn about the advantages of using the PDB-IHM system to make complete depositions of 3DEM/Xlinking-MS-based IHM structures and other types of integrative structures to the PDB.
Structures of complex macromolecular assemblies are increasingly determined using integrative or hybrid methods (IHM), wherein complementary experimental and computational techniques are employed. In addition to traditional structure determination methods such as macromolecular crystallography, NMR spectroscopy, and three-dimensional electron microscopy (3DEM), complementary techniques (e.g., chemical crosslinking-mass spectrometry (Xlinking-MS), small-angle scattering, Förster resonance energy transfer, and chemical shift perturbation without NOE restraints) frequently contribute to integrative structure determination.
PDB-IHM, part of the PDB Core Archive, enables deposition, validation, biocuration, preservation, and dissemination of IHM structures. PDB-IHM infrastructure is supported by the IHMCIF data standard (an extension of the PDBx/mmCIF data standard underpinning the PDB) and software tools that ensure FAIR (findable, accessible, interoperable, and reusable) availability of integrative structural biology data. Importantly, integrative structures processed through PDB-IHM receive PDB accession codes and are archived alongside “traditional” single-method experimental structures in the PDB.
The primary goal of this workshop is to facilitate deposition of IHM structures using PDB-IHM functionality, thereby eliminating barriers to fully archiving these structures in the PDB and preventing data loss. Given the predominance of combined 3DEM/Xlinking-MS-based integrative structures, the workshop will highlight these use cases. Through a combination of short lectures and hands-on practical sessions, participants will learn about (a) IHM data deposition requirements, standards, and practices, (b) pre-deposition file preparation, (c) deposition process and workflows, (d) depositing structures through the web interface, (e) submitting multiple structures using an API, and (f) interpreting the information presented in wwPDB validation reports.
Visit IUCr 2026 for complete details about this structural science meeting.
Depositors should register for the workshops during the registration process; early registration ends March 31, 2026.
01/07/2026
Time-stamped Copies of PDB and EMDB Archives
A snapshot of the PDB Core archive (https://files.wwpdb.org, https://s3.rcsb.org) as of January 01, 2026 has been added to https://s3snapshots.rcsb.org (AWS), snapshots.rcsb.org (rsync -rlpy -a -v --delete snapshots.rcsb.org:: .), and ftp://snapshots.pdbj.org. Snapshots have been archived annually since 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20260101 includes the 246,905 experimentally-determined structure and experimental data available at that time. Atomic coordinate and related metadata are available in PDBx/mmCIF, PDB, and XML file formats. The date and time stamp of each file indicates the last time the file was modified. The snapshot of PDB Core Archive is 1,583 GB.
A snapshot of the EMDB Core archive (ftp://ftp.ebi.ac.uk/pub/databases/emdb/) as of January 01, 2026 can be found in https://ftp.ebi.ac.uk/pub/databases/emdb_vault/20260101/ and ftp://snapshots.pdbj.org/20260101/. The snapshot includes 52,943 3DEM entries. The Snapshot of the EMDB archive is 28.2 TB in size.